Breaking Research from "Microbiome: Therapeutic Implications" With Dr. Elena Verdu
As the “Microbiome: Therapeutic Implications” meeting wraps up in Killarney, Ireland, many hot topics and scientific insights have emerged over the past few days.

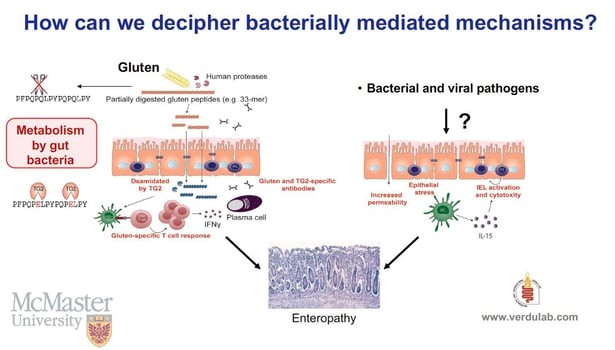
One of the highlights that created a lot of buzz on twitter (#KSmicrobiome), was the presentation by Dr. Elena Verdu, of McMaster University in Ontario, Canada, about how the microbiome influences the development of celiac disease (an autoimmune condition triggered by gluten protein found in wheat and other grains), via metabolism of dietary gluten.
Here we catch up with Dr. Verdu about these revelations regarding the interplay between microbiota, diet and autoimmune disease, and her experiences at this meeting of the minds in Killarney.
Follow meeting highlights and join the conversation on twitter @KeystoneSymp #KSMicrobiome and stay tuned for more blog posts on meeting outcomes.
How do gut-bacteria metabolize gluten?
Dr. Verdu: It has been known for some time that environmental bacteria and fungus produce endopeptidases that degrade gluten immunogenic sequences in vitro. This is the basis for a potential “enzymatic therapy” in celiac disease, which could break down immunogens before the immune system has a chance to react.
We found that the human gastrointestinal tract is colonized by microbes that participate in gluten metabolism in the small intestine. Since gluten proteins are resistant to mammalian enzyme proteolysis, they become an attractive substrate for gut microbial metabolism. We found sequential gluten degradation by eukaryotic enzymes; an opportunistic pathogen producing elastase and core small intestinal commensals leading to various degrees of immunogenic peptide production and detoxification.
How does this contribute to celiac disease in some people but not others?
Dr. Verdu: Our studies identified a specific proteolytic profile in the duodenum of people suffering from celiac disease as compared with non-celiac controls. We also found an increased abundance of an opportunistic pathogen (Pseudomonas aeruginosa) producing elastase-- an enzyme able to cleave some immunogenic sequences in gluten peptides. However, the peptides resulting from this partial, opportunistic pathogen hydrolysis, retained immunogenic capacity, as they were able to stimulate Interferon gamma responses from gluten-specific T cells isolated from celiac patients. These peptides also showed increased barrier translocation compared with native (non-bacterially modified) gluten peptides, such as the 33mer.
Individuals without celiac disease had higher abundance of lactobacillus in the duodenum. In vitro experiments showed that lactobacillus further metabolized the immunogenic peptides produced by the Pseudomonas-elastase activity, leading to very short peptides that failed to activate gluten-specific T cells from celiac patients.
"It is possible that individuals with celiac disease harbour a duodenal microbiota with impaired capacity to detoxify gluten."
~ Dr. Verdu
In people with genetic susceptibility, the balance between small intestinal opportunistic pathogens and commensals could affect the pool of immunogenic gluten peptides produced during normal digestion, their capacity to translocate the gut barrier and modulate celiac disease risk.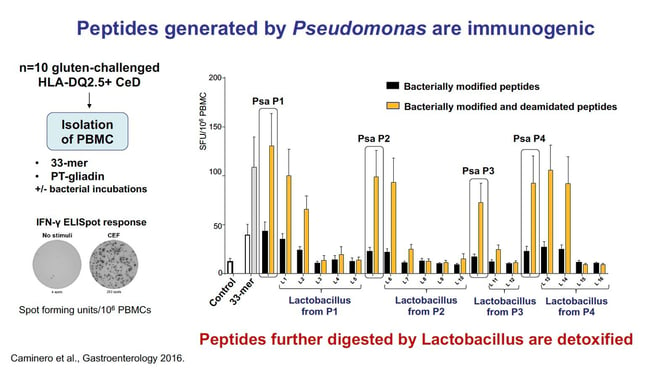
How might these insights about the Microbiome be leveraged to treat or prevent celiac disease?
Dr. Verdu: Many of these steps could be targeted therapeutically and lead to new drug development. For instance, a community of microbes with optimal gluten detoxification capacity could be assembled. From our studies, it seems more likely that sequential degraders are needed for detoxification rather than a single “probiotic” organism.
Pharmacological approaches that target bacterial elastase activity and its direct pro-inflammatory events could also be developed, as well as inhibitors of the receptors involved in its signaling.
Does this work have any relevance to gluten sensitivity or allergy that does not fall under the category of autoimmune disease?
Dr. Verdu: Celiac disease is a very well defined autoimmune and inflammatory disorder. The clinical spectrum of “gluten-related disorders” include conditions that are less well understood such as wheat allergy and non-celiac wheat sensitivity. While gluten is the most important environmental trigger in celiac disease, the antigen/s or exact components in wheat that cause other wheat sensitivities and allergy, remain controversial. However, both gluten proteins and non-gluten proteins in wheat (i.e. amylase trypsin inhibitors or ATI) have been suggested as triggers.
The group of Dr. Schuppan in Mainz has shown that wheat ATI stimulates innate immune responses and we have recently shown that although ATI are proteolytic resistant, they can be degraded by lactobacilli. Thus, the concept of microbial metabolism and detoxification of wheat proteins can also bear relevance to non-celiac wheat sensitivities.
What implications do these discoveries have for other gastrointestinal diseases, or disease beyond the gastrointestinal tract?
Dr. Verdu: Gluten and ATI have been implicated in functional gastrointestinal disorders such as irritable bowel syndrome, and ATI have been implicated in exacerbating experimental colitis. Therefore, these concepts could be applied to IBD, and disorders of the microbiota-gut-brain axis.
Did you make any new connections at the meeting that will help to progress your work or lead it in new directions?
Dr. Verdu: During the first day of the symposium I connected with colleagues and basic microbiologists and had many interesting discussions on the role of microbial inhibitors of elastase activity. I hope these discussions will crystalize into future collaborations!
What did you enjoy most about the Keystone Symposia meeting on the Microbiome?
Dr. Verdu: I mostly enjoyed the friendly, relaxed and collegial atmosphere, and the respectful attitude that allowed all speakers to discuss unpublished data and emerging concepts.
For more information on Dr. Verdu's research, visit her lab webpage:
Recent publications:
- Nat Commun. 2019 Mar 13;10(1):1198. doi: 10.1038/s41467-019-09037-9.
- Gastroenterology. 2019 Jun;156(8):2266-2280. doi: 10.1053/j.gastro.2019.02.028.
- Nat Rev Gastroenterol Hepatol. 2019 Jan;16(1):7-18. doi: 10.1038/s41575-018-0064-z.
- Gastroenterology. 2016 Oct;151(4):670-83. doi: 10.1053/j.gastro.2016.06.041.
For more about the "Microbiota: Therapeutic Implications" meeting, see our:
Q/A blog post with co-organizer Dr. Fergus Shanahan
Couldn’t make it to Killarney? Don’t miss out on upcoming microbiome meetings in 2020
Related news
Keypoint Newsletter: Health Equity Programming
New this year we added new Health Equity Programming to select meetings, to raise awareness for...
Climate Change & Infectious Disease Threats ePanel ON DEMAND
As climate change alters temperatures, precipitation patterns and other habitat features around the...